The Deuteration JRA: capitalising on novel methods for sample production in biological systems
By Trevor Forsyth, Michael Haertlein, Michael Sattler, Luke Clifton, Christine Ebel and Hermann Heumann
This JRA has been developing protocols that are impacting strongly on the scope and quality of biological neutron scattering experiments carried out at central facilities throughout Europe. The need for this is very clear given the increasing trend towards interdisciplinary and integrated approaches for the study of biological systems. Neutron scattering has a unique and important role to play if the right types of sample can be made available. Deuteration is essential to this: the ability to label complex/interacting systems offers approaches that are simply not possible using other methods. This project is widening the access of neutron scattering methods to biologists throughout the European Union, both by extending the range of problems that can be tackled, and by reducing the cost impact of sample preparations. It exploits an obvious synergy with the nuclear magnetic resonance (NMR) community, which also has important needs for isotope labelling and within which there is increasing use of neutron scattering.
Optimisation of deuterated biomass production
A large number of biological neutron studies rely on the biosynthesis of deuterated biological macromolecules using deuterated carbon sources. The cost of these often limits experiments severely. In this task protocols have been developed for the optimisation of E. coli and algal biomass production. Since algae grow photosynthetically, relying only on D2O medium, CO2, and light, they are an ideal candidate for cheaper production of deuterated components. Major downstream applications in neutron scattering are obvious for these in vivo products: deuterated molecules can be purifi ed and used as deuterated feedstock to bacteria such as E. coli.
Production of labelled proteins in yeast
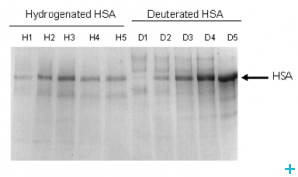
Most proteins used in neutron scattering studies are produced in bacteria such as E. coli. This type of protein production has had a huge impact on studies of biological systems using a wide variety of techniques. In many cases the use of E. coli is limited by folding/post translational modification problems. Some proteins have to be produced in lower eukaryotic expression systems. However there are difficulties in adapting such cells to growth in deuterated media.
Here we have developed methods whereby labelled proteins can be produced (intracellularly or exported) in yeast.
Segmental labelling
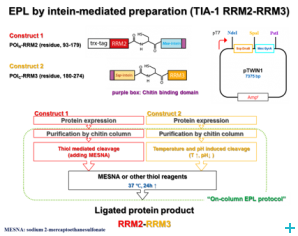
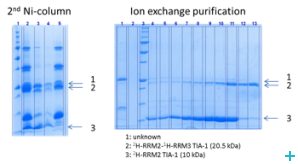
Over the course of the project we have established two model systems (U2AF65 and TIA-1) for implementing and optimizing protein ligation techniques and subsequently demonstrated the expression of fragments and the ligation using protein transsplicing techniques. The production of segmentally isotope-labeled proteins was first established using segmentally 15N-labeled proteins. The optimized protocol was then used for segmental deuteration of the protein TIA-1 RRM2-RRM3. The purified protein was used for SANS measurements with contrast variations at Institut Laue-Langevin (ILL) Grenoble to document the utility of segmental deuteration.
Based on the results obtained in this project a protocol for the production of segmentally deuterated proteins has been developed, which serves as a guideline and recommendation for the neutron community. The experimental protocols and optimisations of the production of segmentally deuterated proteins have been exchanged with the Life Sciences group at the ILL and will thus be available for the general scientifi c community of neutron researchers. The implementation of the protocol is being prepared for publication.
We have also started to explore the use of sortase-mediated protein ligation, which has been reported to yield better ligation efficiencies compared to intein-based methods. For the TIA-1 model systems, this work is still in progress. However, for another multi-domain protein we have successfully implemented sortase-mediated protein ligation with very high yields.
Low cost D-glycerol production
The high cost of deuterated carbon sources for the expression of D-proteins in E.coli is still a stumbling block for the application of neutrons in biology. In this task, methods have been developed for the production of deuterated glycerol by algae under salinic stress. A novel protocol based on “milking” algae of the glycerol produced, dramatically reduces costs since the same biomass and D2O can be used repeatedly. Approaches have been developed using Dunaliella salina and Chlamydomonas reinhardtii (a freshwater algae that could be competitive with Dunaliella). Dunaliella accumulates up to 6M glycerol in the cell and this task has focused on efficient release of glycerol into the media. Methods tested included high temperature treatment, cell immobilization and mild sonication.
Deuterated membrane proteins
Membrane proteins perform a wide range of essential cellular functions and play key role in (for example) transportation, energy management, signal transduction, photosynthesis. They are also implicated in a number of genetic diseases and have considerable therapeutic importance (70% of drug targets). This group has focused on the development of methods to optimise deuteration of membrane proteins. Model membrane proteins have been identified and bacterial expression systems in high cell density cultures used for deuteration.
The deuterated membrane proteins will be used to reconstitute membrane systems with hydrogenated lipids for neutron studies and to test new classes of surfactants for their capacity to stabilize functional assemblies.
Deuterated lipids
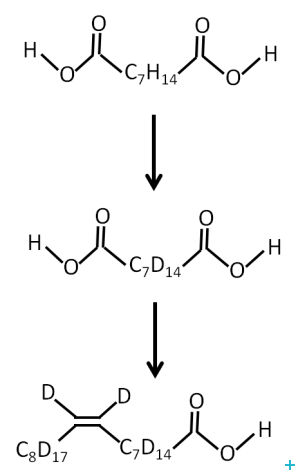
One of the major limitations for the application of neutrons to the study of biological systems is the availability of deuterated lipids. Membrane biology is a particularly important area and a new range of powerful experiments could be carried out with the availability of selectively labelled lipids. The provision of unsaturated perdeuterated lipids with a range of head groups required the development of an optimised route for the production of oleic acid and conversion to the target lipids.
We have developed a methodology for the production of full and part deuterated oleic acid. Coupling of the acids to phosphocholine head group is straightforward chemistry that is accessible to a wide range of academic and commercial groups.
We plan to pass the material to a range of partners to both spread the load and develop a wider range of sources for the produced material. The phosphoglycerols (and phosphoserines) are synthetically accessible from the phosphocholines so these will be the main focus.
In conclusion, an optimised route for the production of fully and part deuterated oleic acid was developed. Stockpiling of this material should enable milligram quantities of deuterated unsaturated phospholipids for use in neutron scattering experiments to be made available at relatively low cost.
Conclusions and future work
The tasks carried out as part of this JRA have provided new capabilities in the area of biological neutron scattering and this is having a significant impact in widening capabilities throughout this part of the user community. For the future there are a number of key areas where further method development will be focused. For example we believe that major progress is possible for the culture of mammalian cells, cell-free synthesis, and in a number of key scientifi c areas such as the study of intrinsically disordered proteins (IDPs).
In the case of mammalian cell systems, the drive is to allow the biosynthesis of deuterated macromolecules that cannot be obtained using bacteria, yeast, or algae. This is essentially unexplored territory, and will need careful development of adaptation processes. Success in this area will have an enormous impact on availability of molecules that cannot be obtained by other recombinant methods. A good example is cholesterol – currently impossible to obtain in perdeuterated form because chemical synthesis is too complex and biosynthesis is impossible in bacteria.
Cell-free systems are also of strong interest; these can be used for the production of recombinant proteins, and have several advantages over traditional in vivo expression. For neutron scattering, a cell-free method developed and optimised for deuteration will allow amino acid labelling without isotope scrambling amongst the amino acids within the protein, and the production of toxic labelled proteins. The exploitation of the technique has been limited in the past by low yields. However the technology has changed and the approach is now feasible. A successful initiative in this area would also allow multiple selective amino acid labelling – where in vivo methods are very complex.
Deuterated intrinsically disordered proteins (IDPs) are also of central interest. IDPs are an important class of proteins that are either fully unfolded or contain significant contiguous regions that are devoid of secondary structural elements. IDPs play key roles in a vast range of physiological processes, and are also strongly implicated in important human diseases such as cancer and neurodegenerative diseases like Alzheimer’s disease and prion-related diseases. The heterogeneity of these proteins means that crystallography is appropriate for their study. Selective deuteration will allow SANS and incoherent scattering studies in which there is a specific focus on a particular part of the protein. Method development will need to focus IDP systems where the component parts are expressed separately in E.coli together with intein sequences and then ligated in vitro – i.e. exploiting the types of segmental labelling that have been developed during the period of this JRA.
- Trevor Forsyth, JRA coordinator, Institut Laue-Langevin (ILL), Grenoble, France & EPSAM/ISTM, Keele University, UK
- Michael Haertlein, ILL
- Michael Sattler, Technische Universität München (TUM), Garching, Germany
- Luke Clifton, SFTC Rutherford-Appleton Laboratory, Didcot, UK
- Christine Ebel, Institut de Biologie Structurale (IBS), Grenoble, France
- Hermann Heumann, Max Planck Institute (MPI), Martinsried, Germany
Relevant references:
- Abla M. et al. (2012) J. Fluorine Chem., 134, 63-71.
- Blesneac I. et al. (2012) Biochim. Biophys. Acta 1818, 798-805.
- Cuypers M. et al. (2012, in press) Angew. Chem.
- Gallat F.-X. et al. (2012) Biophys. J., 103, 129–136.
- Grage S., et al. (2011) Biophys. J., 100(5) 1252–1260.
- Kennaway C. et al. (2012), Gene Dev., 26, 92-104D.